
Selected Projects
[TransMed, NGS] Metagenomic sample phenotype prediction using machine/deep learning
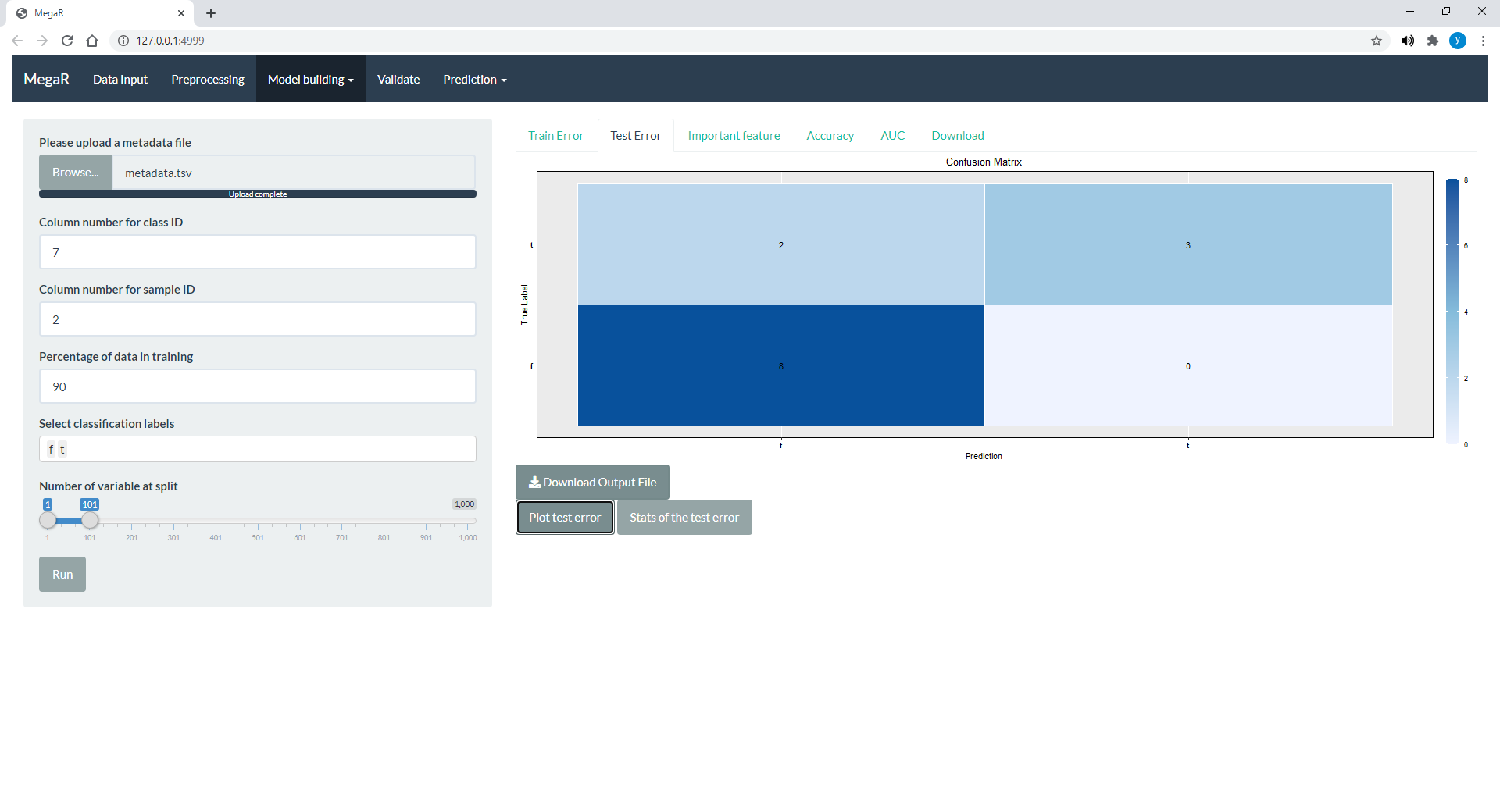
Project Summary: Machine learning has been utilized in many applications from biomedical imaging to business analytics. Machine learning is stipulated to be a strong method for diagnostics and even for determining therapeutics in future as we move to personalized medicine. MegaR provides an unprecedented opportunity to develop machine learning models from metagenomic data available publicly as well as to perform classification of data samples based on the optimal model we developed.
Skills: R, R-shiny, Machine Learning (Scikit-Learn), Deep Learning (Keras, TensorFlow)
Paper: [BMC Bioinfo 2021],
Poster: [ACM-BCB 2020]
[TransMed, NGS] Immune T-cell analysis using machine/deep learning
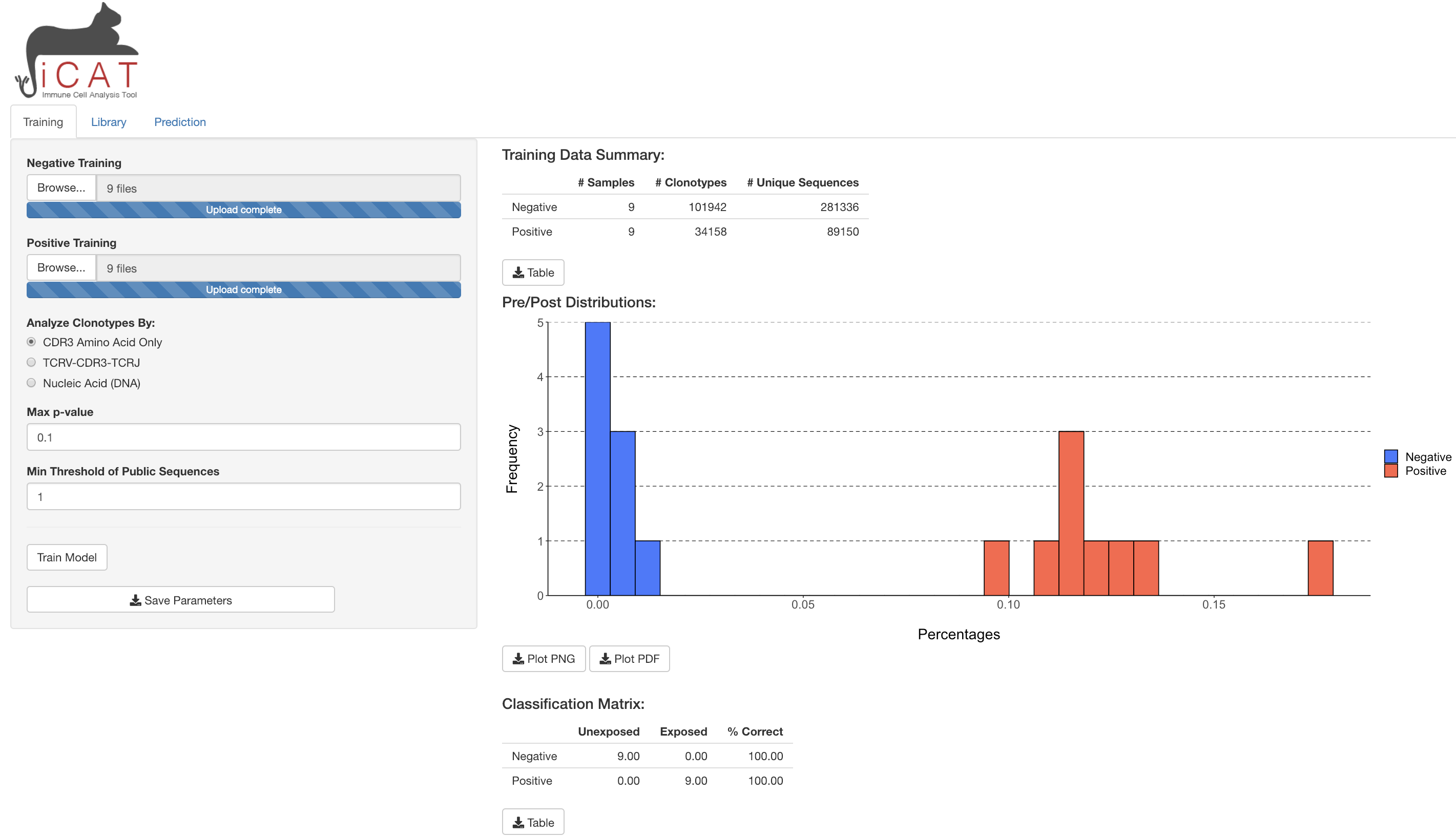
Project Summary: T-cells are vital to the adaptive immune system, recognizing pathogens through the T-cell receptor (TCR).
For each T-cell, the TCR loci undergoes genetic rearrangement which can act as a unique biomarker, cataloging immunological history.
iCAT is a user-friendly, graphical-interface software that analyses TCR-specific sequencing data from exposed (positive) or unexposed (negative) individuals to identify TCR sequences statistically associated with positive but not negative samples.
We are recently developing a novel method to analyze complex and large scale human datase using deep learning techniques.
Skills: R, R-shiny, Machine Learning (Scikit-Learn), Deep Learning (Keras)
Collaborator: Dr. Richard DiPaolo (Saint Louis Univ. School of Medicine)
Paper: [F1000 Research 2021],
[ACM-BCB 2020],
[Cell Reports 2018]
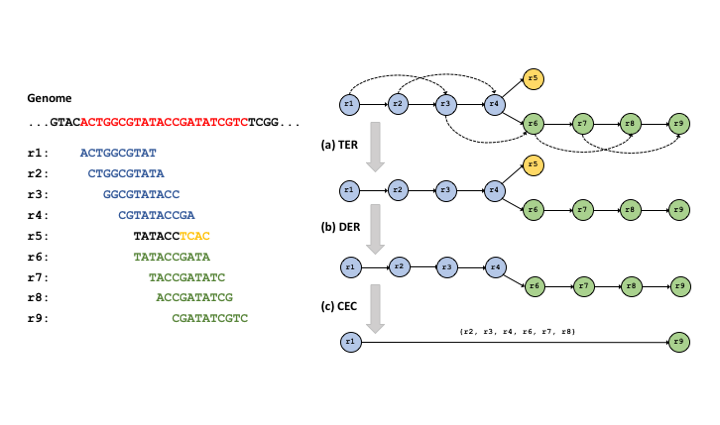
[HPC, NGS] Apache Spark: Scalable Overlap-graph Reduction Algorithms for Genome Assembly in the Cloud
Project Summary: The advent of high-throughput DNA sequencing techniques (next-generation sequencing) has permitted very high quality de novo assemblies of genomes, but raise an issue of requiring large amounts of computer memory to resolve the large genome graphs.
To address these limitations, we present a novel algorithmic approach; Scalable Overlap-graph Reduction Algorithms (SORA).
SORA adapts string graph reduction algorithms for the genome assembly using a distributed computing platform.
The experimental results show that SORA can process a nearly one billion edge graph in a distributed cloud cluster as well as smaller graphs on a local cluster with a short turnaround time.
Skills: Apache Spark, GraphX, Amazon Cloud, Scala, Python, Shell
Paper:
[Human Genomics 2019],
[IEEE-BMBM 2018],
Poster:
[ACM-BCB 2017]
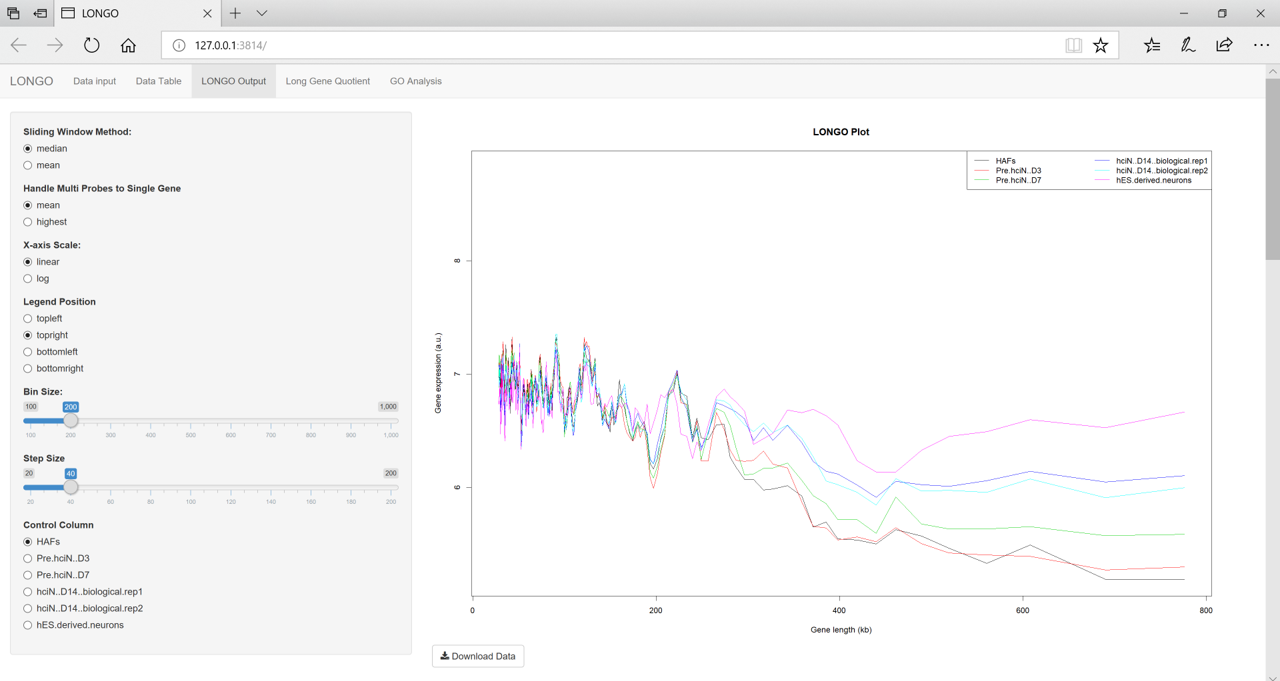
[TransMed] R-package: Gene Length Depenent Analysis for Neuronal Conversion
Project Summary: Gene Length-Dependent Expression Analysis Tool in Neuronal Cells
Skills: R, R-shiny, BioMart, GO analysis
Collaborator: Dr. Andrew Yoo, Washington Univ. School of Medicine
Paper: [Bioinformatics 2018]
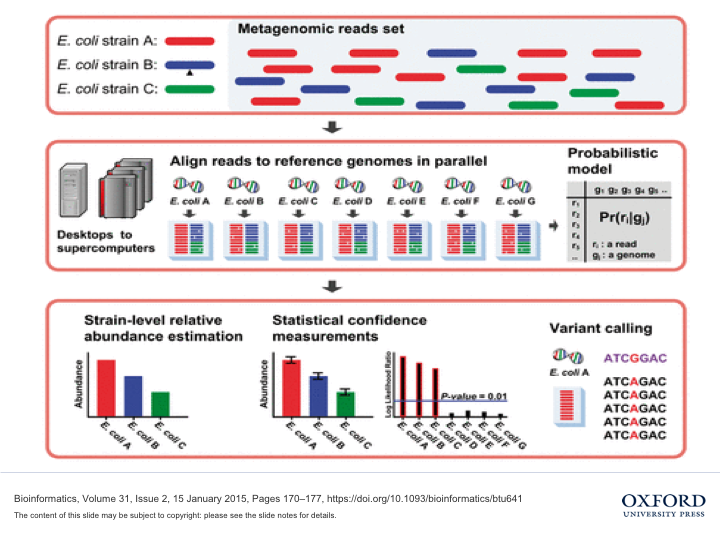
[Metagenomics, NGS] Metagenomics Analysis: Strain-Level Taxonomy Identificaiton Tool, Metagenome Assembler
Project Summary: Strain-level genome identification algorithm for biosurveillance using high-performance computing
Skills: C++, Python, MPI, OpenMP, Supercomputer
Paper:
[Bioinformatics 2015]
Project Summary: An overlap-graph de novo metagenome assembler
Skills: C++, Python
Paper:
[Bioinformatics 2014]
Ph.D Research (until 2012)
[CompBio] Cell Cycle Modeling and Stochastic Simulation using HPC
Project: Implicit Stochastic Simulation Algorithm for Chemical Kinetics
Skills: Fortran, Java, C++, MPI, OpenMP, Supercomputer
Papers: [JAAC 2015], [IJPP 2015], [ICCS 2011], [ACM-BCB 2010]
Skills: Fortran, Java, C++, MPI, OpenMP, Supercomputer
Papers: [Cell Cycle 2011], [CMES 2009], [SpringSim 2009]
[HPC] Internship projects
Skills: MATLAB, MATLAB Parallel Computing Toolbox, Distributed Computing Server
Skills: C++, Python, MPI, OpenMP, Supercomputer
Papers: [IEEE Access 2013], [SIMULTECH 2011]