Lab
|
Topic:
|
Phylogenetic Tree Visualizations
|
|
Collaboration Policy:
|
The lab should be completed working in pairs
|
|
Submission Deadline:
|
|
Contents:
Overview
Our textbook authors prepared a series of labs associated with
Chapters 9,10 of the text (part1,
part2,
part3,
part4),
and while you are welcome to go through those labs on your own time,
there is something dissatisfying about the drawing algorithm that they
suggest. While they look nice on the examples they use, such as with
the problem is that with larger trees, their algorithm does not
disallow portions of one branch of a tree from overlapping the
visualization of another branch of the tree. For example, applying
their algorithm to a more complex tree produces the following image:

While their algorithm, with sloped lines, could be remedied, we will
instead explore a different visualization style that uses orthogonal
lines, producing images such as the following for the above examples:


For full disclosure, this example is modeled from an example in a 2011 article
Megacycles of atmospheric carbon dioxide concentration correlate with
fossil plant genome size.

Files You Need
We are providing you with two files:
-
treeViz.py
We have provided some utility functions to get you started,
and we have stubbed out the draw1 and draw2
functions which you are to implement.
-
samples.py
This file provides a variety of sample trees for use in
testing your software (many of which are described below as examples).
Your Tasks
There will be three requirements for your submission of this lab, each
of which is described in far more detail in the remainder of this document.
-
Warmup questions
Before doing any coding, we need you to better understand the
recursive nature of the drawing algorithm, so we will ask you to
do some pen-and-paper examples (but to later type up your
answers within comments of your source code).
-
draw1 function implementation
Then you will implement the first version of the drawing algorithm.
-
draw2 function implementation
We will guide you through a more advanced refinement of the
algorithm that varies edge lengths to match presumed
evolutionary time period.
Turtle Graphics Primer
The turtle module is part of Python's standard libraries, to
provide an illustrative way to generate some basic graphics. The
turtle is effectively a virtual robot with a pen that draws lines as
it moves. The only behaviors you will need to use are:
- turtle.forward(distance)
- turtle.backward(distance)
- turtle.left(degrees)
- turtle.right(degrees)
- turtle.write(message)
For convenience in testing, we have provided a function
reset() within our code that clears the screen and returns
the turtle to a starting position near the left edge of the
screen. You should call reset() just before starting your
draw function (but not from within!).
Algorithm Overview
Our drawings will be produced with a recursive algorithm. By
convention, we will assure that when drawing a tree, the turtle begins
facing rightward at the point that should become the left edge of the
tree visualization, and with a vertical position that should be the
vertical center of the eventual tree. Furthermore, we will assure
that the leaves of the tree are to be laid out vertically at
regular intervals (which we will denote as yScale within our
later functions). For example, we might decide that leaves will be
drawn at 20-pixel intervals on the vetical scale. By this convention,
a generic schematic of a tree with eight leaves might appear as
follows:

with the solid rectangle meant to portray the bounding-box of the
tree, and the eight dashed horizontal lines designating the vertical
location of where those eight leaves will eventually be drawn. Notice
that the turtle starts precisely at the vertical center of the image.
With a recursive approach, the key insight is that if we have a tree
that is a single leaf, we simply need to write the text information
about the leaf. For any other tree, rather than worrying about all
the complexity of the tree, we want to do the following basic steps:
-
Turn to move upward, then rightward, leaving the turtle at the
appropriate starting spot for the first subtree.
-
Recursively draw that subtree (which must leave the turtle
precisely located and oriented where it started).
-
Retrace the path to the root and then move downward,
then rightward, leaving the turtle at the
appropriate starting spot for the second subtree.
-
Recursively draw that subtree (which must leave the turtle
precisely located and oriented where it started).
-
Retrace the path to the root and leave the turtle oriented
facing rightward (just as we found it when beginning this
process).
The key to the success of our algorithm (and avoiding overlap of
subtrees) is in determining precisely how far
upward/downward/rightward to move before restarting each recursive
drawing. In determining how far upward/downward to move, we must rely
on our convention that the eventual leaves of the tree be evenly
spaced on the vertical scale. If we knew how many leaves were in the
first and second subtree, we should be able to determine the correct
vertical offsets.
As a first example, consider the generic 8-leaf tree and presume that
we knew that the first subtree had 3 of those leaves and the second
subtree had 5 of those leaves. In this case, we should envision the
recursive process as follows:

Notice that the top of the two subtree bounding boxes will cover three
of the eventual leaves and we will bring the turtle precisely to the
center of the left edge of that bounding box before starting the
recursion. The bottom of the two subtrees will have five leaves, and
thus we can determine where the left-center of that box should be.
Of course, if we had a different distibution of leaves, we would need
our algorithm to "deliver" the turtle to other locations. For example,
here is a schematic for an 8-leaf tree with 6 leaves in the first
subtree and 2 in the second.

For convenience, the Python code we are providing you with already has
a function with signture leafCount(tree) that returns the
number of leaves within a given tree or subtree.
Warmup Questions
This brings us to your first part of the lab. We must eventually come
up with a programatic way to determine the various distances for a new
tree that we encounter. But before getting bogged down in Python code,
you need to work out some cases by hand, and then hopefully determine
a pattern that will allow you to generalize this to arbitrary size trees.
The question at hand is if we assume that the turtle starts at
coordinate y=0, we wish to determine what the y value
should be for starting the first subtree and what the
y value should be for starting the second subtree. For
the sake of these examples, let's assume that the leaves are rendered
10 pixels apart from each other. (In our real code, we'll make that
yScale a parameter.)
Also, unlike mathemticians, computer scientists tend to count pixels
from the top of the screen downward, and thus we consider moving
upward in the negative direction and downward in the
positive direction.
Revisiting our first example of an 8-leaf tree with 5 leaves in
its first subtree and 3 in its other, the first subtree should begin
at height y=-25 and the second at height y=+15.
In the second example, with 6 leaves in the top and 2 in the bottom,
the starting heights for the recursions were y=-10 and
y=+30 respectively.
You must complete the following table (which we've placed within the
comments of the source code we are providing).
total
leaves |
upper
leaves |
lower
leaves |
upper
y-value |
lower
y-value |
| 8 |
3 |
5 |
-25 |
+15 |
| 8 |
6 |
2 |
-10 |
+30 |
| 8 |
4 |
4 |
|
|
| 8 |
7 |
1 |
|
|
| 7 |
3 |
4 |
|
|
| 7 |
1 |
6 |
|
|
| n |
a |
b |
|
|
Note well that the last entry of this table is the most important, as
you need to discover a formula that works for general parameters
(presuming that a+b=n).
draw1 function
Now we are ready to write some code. We will do two versions of the
visualization that differ in how they manage the horizontal
spacing of the drawing. In the first version, we will simply
move a fixed amount rightward for each level of the tree. The function
should have signature
def draw1(tree, xScale, yScale):
where xScale defines that horizontal offset per level, and
yScale is the vertical offset from leaf to leaf.
The code for your implementation should follow the high-level
algorithm enumerated above, distinguishing
between a base case where you have a tree with empty subtrees and the
general case in which the subtrees are nontrivial. The above turtle graphics primer can guide you through use of
the graphics package.
We have included a variety of sample trees within the Python
code. Here are some renderings for you to match:
Tree from Figure 9.3 of our book, rendered as
draw1(fig93, 50, 50):

Tree from Figure 9.4 of our book, rendered as
draw1(fig94, 50, 50):

Rendered as draw1(treeFrogs, 50, 50):

Rendered as draw1(complex, 40, 15):

draw2 function
The difference between the draw1 and draw2 functions
involved the rightward spans of the edges when moving from a branch
point to its subtrees. With draw1, we
simply used a fixed increment for each rightward movement.
However, leaves of phylogetic trees are often based on
relatively modern day samples of organisms, while internal nodes
represent hypothesized common ancestors. Therefore, visualizations
that one to capture the history align all of the leaves at the far
right of the figure, and internal nodes can be augmented with a
numeric value that estimates how long ago that common ancesstor
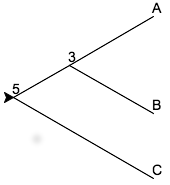
branched. For example, in the following tree (named withLengths in the samples)

we might presume that the 3 denoted at the nearest ancestor to A and B
suggests that existed 3 million years ago while the 5 denoting the
common ancestor of that node and C occurred 5 million years ago.
Internally, this tree is represented as follows.
(5,
(3,
("A", (), ()),
("B", (), ())
),
("C", (), ())
)
In the second version of our visualization, named draw2, we
interpret the numbers at those internal nodes as ages, and
modify the lengths of the horizontal edges to reflect the time scale,
such that modern-day is thought of as time 0 at the far right and then
other ancestors are separated based on the time gaps from the
data. For example, the above tree would be rendered in our new format as

The length of the edges from A and B to their ancestor is equal to
three units (times some arbitrary xScale factor that can be
given as a parameter to convert to pixels), and the line from that
ancestor back to the root is length two units (because that connects
the ancestor that was modeled as 3 million years ago to the one that
was 5 million years ago). More generally, when going from a
parent to a child in the tree, the length of the edge should be
proportional to the numeric "age" of the parent and the numeric "age"
of the child (with leaves implicitly having age 0). The
xScale parameter should not be a multiplier to the horizontal length.
Here are a few other examples that are included in our sample data
sets. There is a data set about tree frogs from the textbook authors.
Its internal numbers are shown on this rendering from
draw2(treeFrogs, 50, 50):

Its new rendering as draw2(treeFrogs, 5, 50) appears as:

Finally, here was our rendering of the most complex tree in our data
set, which we rendered with parameters
draw2(complex, 1, 15):

Submitting Your Assignment
One member of your partnership should electronically
submit your modified file treeViz.py. The
comments at the beginning of the file should clearly identify the
member(s) of the partnernship and should include answers to the
"warmup" questions.
Grading Standards
The assignment is worth 10 points, which will be assessed as follows:
-
(3 points)
Correct answers to warmup questions.
-
(5 points)
Correct implementation of draw1 function.
-
(2 points)
Correct implementation of draw2 function.
Note well that the more advanced draw2 is worth a
relatively small percentage, not because it is easy, but so that you
are able to get 8/10 points just by correctly completing the warmup
and the first implementation correctly.
Last modified: Monday, 26 March 2018













